Table of Contents
Quick Notes
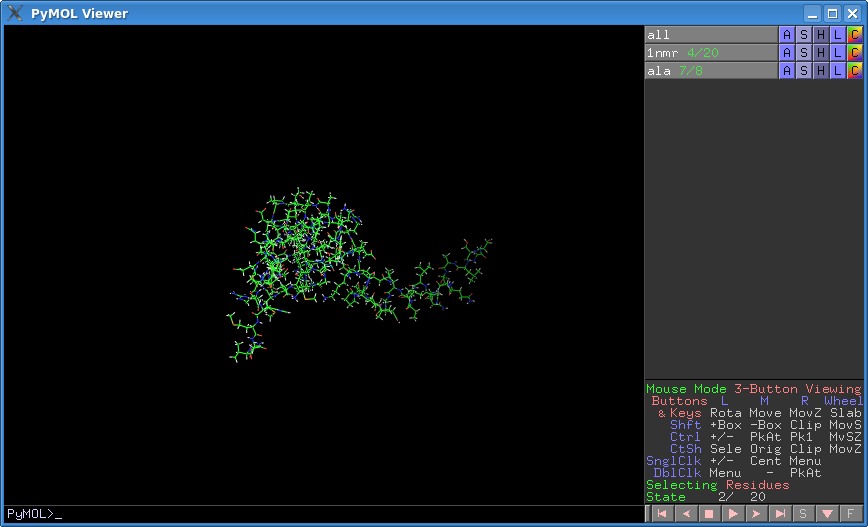
- PyMOL now shows each object's state counter by default. This is useful when measuring from two different states in two different objects.
- You can turn the state counter on or off.
- You can set the state counter display mode to show the current state over all states, eg. 3/17, or just the state number, eg 3.
- Tip: If the command line is empty, use the right and left arrow keys to cycle through states.
States in PyMOL
PyMOL has two types of states. The first is called the 'scene' or 'global' state. By default, all objects are set to match this 'global' state number, or they default to state 1 if they only have one state. The second type of state is an object state. This type of state is independent of the global state and can be set per-object by the user. That is, we can 'freeze' an object into a state by choosing A > Action > State > Freeze, or by typing set state, X, objName where X is the state number and objName is the name of the object.
It is common practice to freeze an object state into state X, and then adjust the global state to something else.
New State Counter
This new state_counter makes it easier to see which state each object is in.
Settings
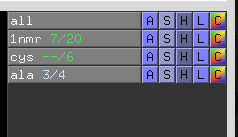
state_counter- 0, off, no object-state counting
- 1, on, on default, fraction-based state counting (eg. 3/4)
- 2, on, state-based state counting (eg. 3)
Examples
- The new state counter appears after the object name. If the global state has gone beyond the object's state, you see “–”; if the object has been frozen, it's colored blue.
- Can 'freeze' to a different state than the 'scene' state}
- Can unset the state_counter to return to normal mode
- State_counter_mode, set two '2' will show just the state
- Can have multiple objects in different states
Improved State Awareness
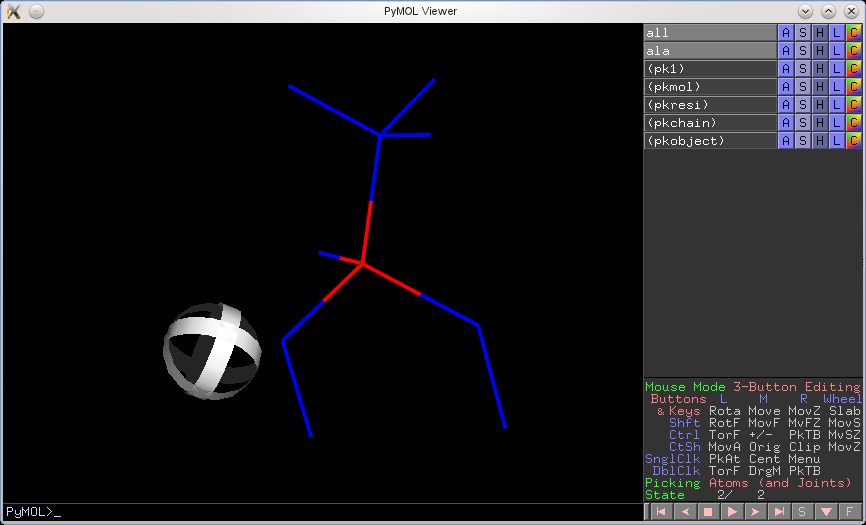
In previous versions of PyMOL, if you were focused on an object in state X, but the global state counter was in state Y, then the visualization and selection mechanism often disagreed. That is, you would click on an atom, and PyMOL would select that atom, but from a different state. The following figure illustrates this behavior. I created a two-state alanine fragment. I then 'froze' the alanine in state 1 and selected the red-colored alpha carbon. PyMOL selected the alpha carbon, but from state 2–not what I wanted.
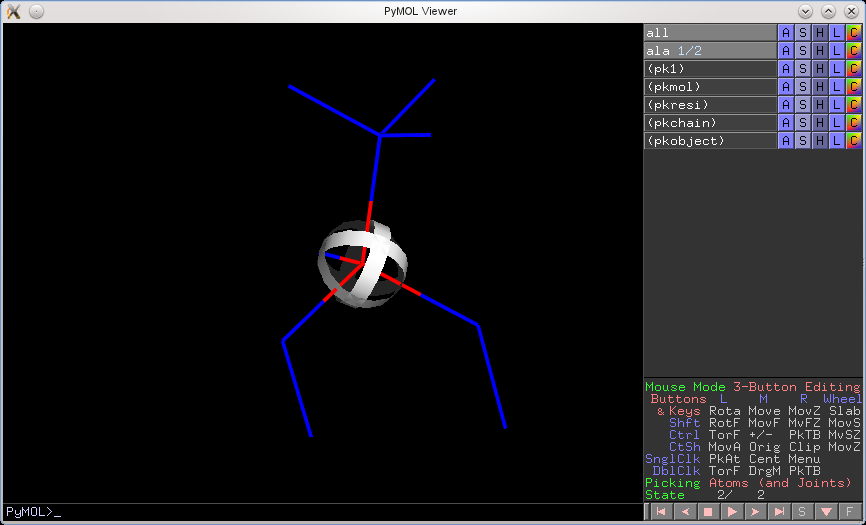
Starting from version v1.4.1, PyMOL will now prefer to select and pick objects in their frozen state, before honoring the global state. Here's the same example as above, but from the improved version of PyMOL.
The same works for selection indicators (the square purplish boxes that appear after you click on something when in Viewing Mode).
Improved Cross-State Object Measurements
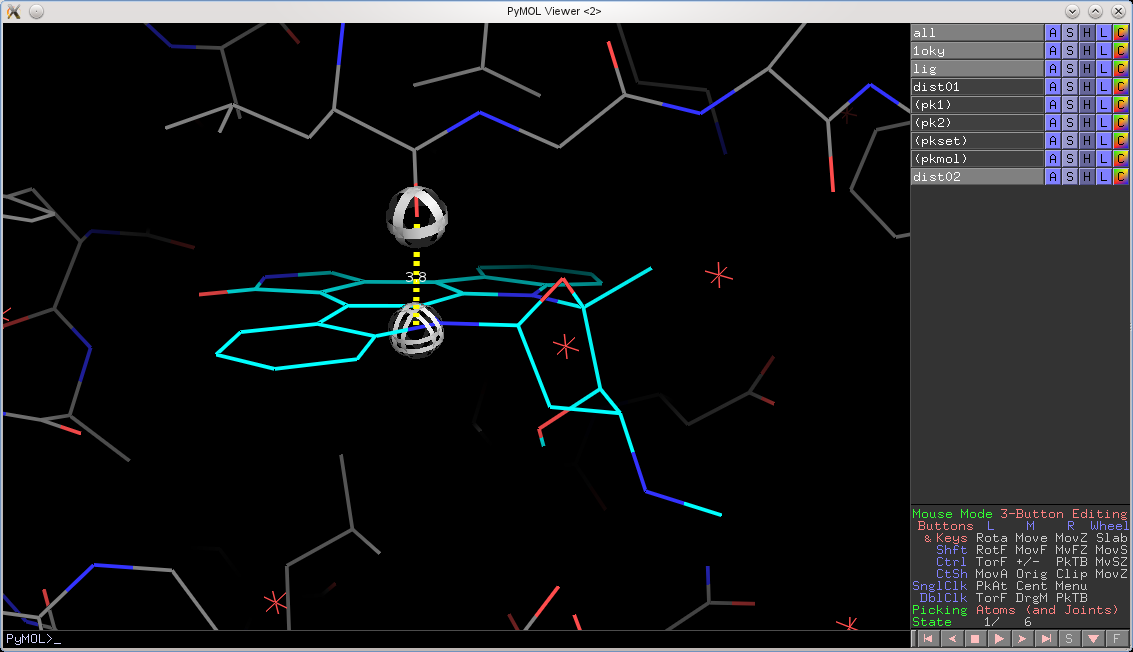
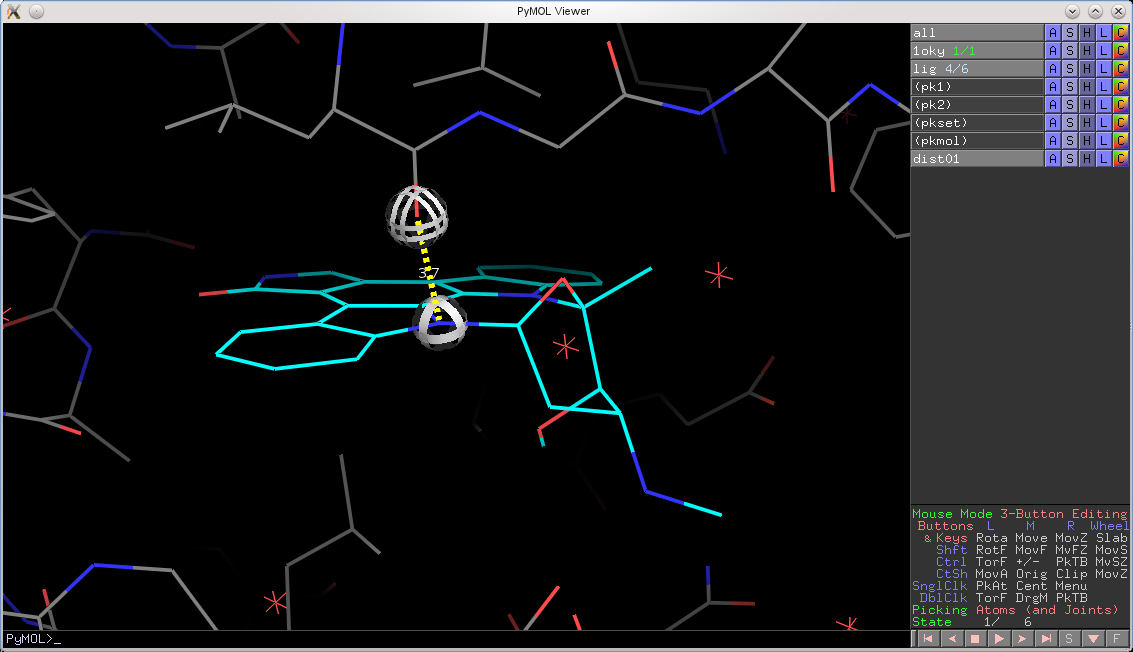
PyMOL will now also honor an object's set state over the global state. For example, if you have docking results in the form of a protein with 30 best poses saved, you can now measure distances, angles, and dihedrals from any state in the small molecule to any state in the protein. Previously, PyMOL would only measure from state X to state X, now it can do any state X to any other state Y.
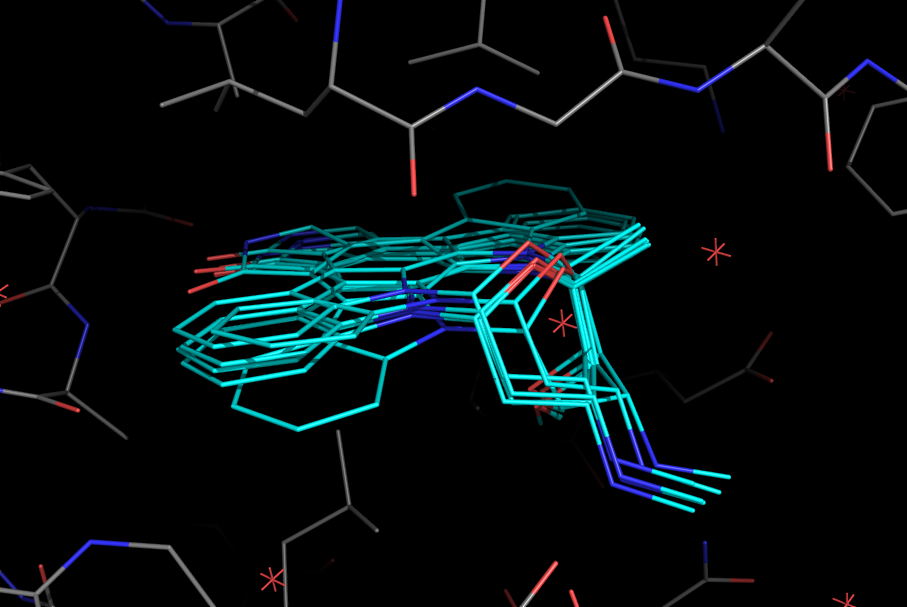
Let's take a look at an example. I setup a protein with a small molecule bound in the pocket over six poses. When all_states are shown, we see all poses:
Now, let's try to measure from state 4 in the ligand to state 1 in the protein using the previous version of PyMOL. We would get the following:
In the new version, the globes hover over the correct locations and the distance is measured between the correct atoms: