Hemoglobin
If you are connected directly to the internet, you can download a hemoglobin PDB structure by simply typing
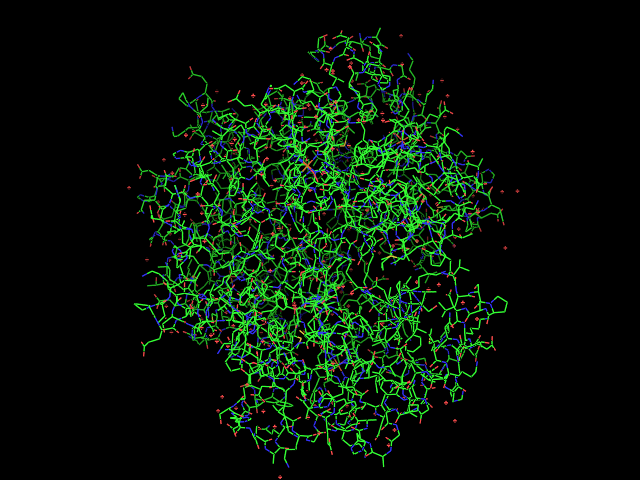
fetch 1gzx
into the PyMOL command line. You screen should then look something like this:
Alternatively, you can download the PDB file from Protein Data Bank and load it manually.
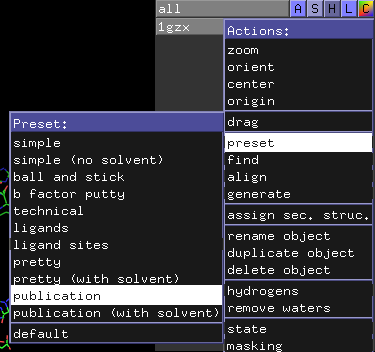
Next, click on the blue “A” button next to the “1gzx” entry on the upper-right-hand area of the viewer window. This should cause the “Action” pop-up menu to appear. Move the mouse down to “preset” and then click and release on “publication”.
Your screen should then look something like this:
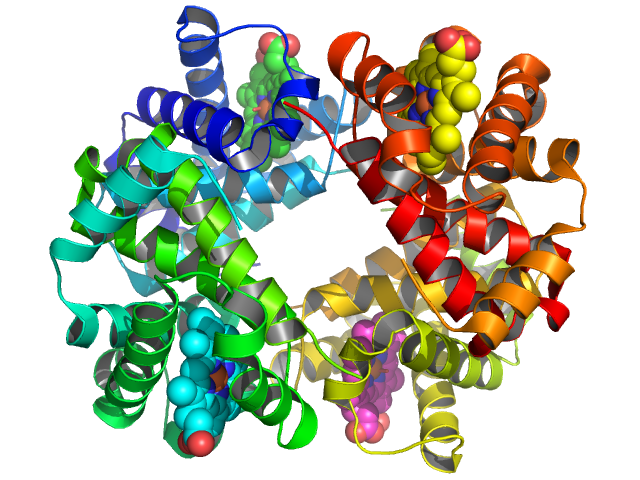
You can manipulate the view manually by left-clicking-and-dragging the mouse in the display area, but you can also “orient” the camera (in the “Action” menu) to get a view showing hemoglobin symmetry.
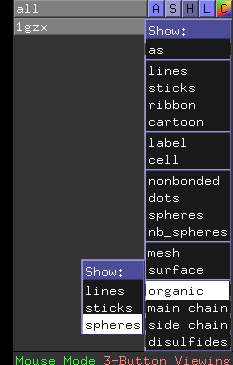
You can zoom in a bit more by right-clicking-and-dragging the mouse downward in the display area, and you can show the four hemes as spheres by clicking on the “S” next to “1gzx” to activate the “Show” menu, followed by “organic” and “spheres”.
to give:
To select a white background, go to the “Display” menu in the upper window and choose “Background” and then “White”.
To eliminate jagged edges, include shadows, and improve the lighting, click the “Ray” button in the upper window area and wait a minute or so in order to get something like this:
You can then save the image to a file by going to the “File” menu and selecting “Save Image” and “PNG…”. On some platforms, you may also be able to select “Copy Image to Clipboard” from the “Edit” menu which can then be pasted directly into Word, PowerPoint, or Keynote, etc.
Congratulations on creating your first PyMOL figure!