loading
Table of Contents
Loading Molecules
Formats
At the present time, Open-Source PyMOL has support for the following molecular formats:
- PDB format with support for various extensions for specifying connectivity, bond valences, multiple models, ensembles, and trajectories.
- MOL format for loading of individual small molecule structures.
- SDF format for loading of multiple small molecule structures.
- MOL2 format for loading systems containing both small molecules and proteins.
- XYZ variants, including multimodel XYZ.
PyMOL also has limited support for
- ChemDraw3D CC1 and CC2.
- The old Macromodel MMD/MMOD format.
- Amber and Gromacs trajectory files, through the VMD plugin architecture.
Incentive PyMOL Builds also support the following proprietary formats:
- MOE Molecular Operating Environment (Chemical Computing Group)
- MAE Maestro (Schrödinger, LLC)
Methods
Molecular structures can be loaded into PyMOL in the following ways:
- via Menus
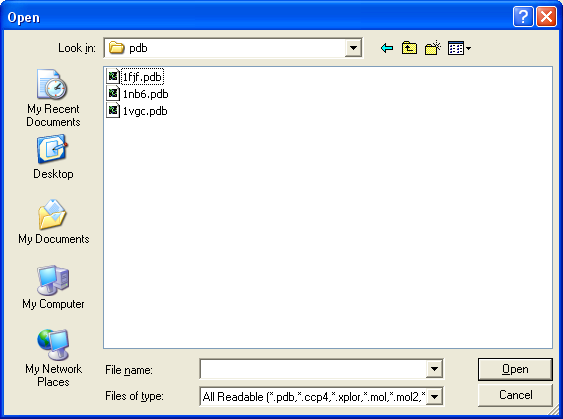
- File Menu: Open
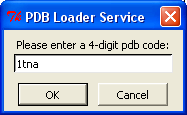
- Plugin Menu: PDB Loader Service
See Also
loading.txt · Last modified: 2013/08/19 21:00 (external edit)