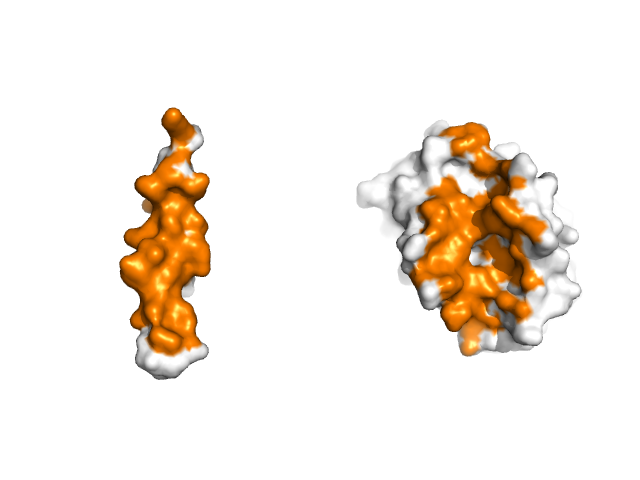figure:splay_interface
Splaying Apart an Interface
The script below generates the figure above. Given the automatic behavior of the script, you should be able to simply update the PDB code and atom selections in order to splay apart the interfaces of other proteins.
# splay.pml - automatically splay apart an interface
# load PDB and split it up into two objects: side1 & side2
fetch 2bzw, tmp, async=0
# in 2bzw, chain A is BCL-XL and chain B is the BAD peptide
extract side1, A//
set_title side1, 1, BCL-XL
extract side2, B//
set_title side2, 1, BAD-PEPT
dele tmp
# define an interface selection
select interf, (side1 w. 4.5 of side2) or (side2 w. 4.5 of side1)
# orient the interface region perpendicular to the camera
# with the long axis horizontal
orient interf
# now turn it so that the long axis is vertical
turn z,90
# make sure side2 atoms are nearer to the camera (UGLY HACK)
_=stored
_.zz = [cmd.get_view()[2], cmd.get_view()[5], cmd.get_view()[8]]
_.z1 = 0.0
_.z2 = 0.0
cmd.iterate_state(1,"interf & side1","_.z1 = _.z1 + x * _.zz[0] + y * _.zz[1] + z * _.zz[2]")
cmd.iterate_state(1,"interf & side2","_.z2 = _.z2 + x * _.zz[0] + y * _.zz[1] + z * _.zz[2]")
z1 = _.z1 / cmd.count_atoms("interf & side1")
z2 = _.z2 / cmd.count_atoms("interf & side2")
del _
if z1>z2: cmd.turn("y",180)
# compute the approximate window width in Angstroms (UGLY HACK)
height_width = cmd.get_session()["main"][0:2]
asp_ratio = float(height_width[0])/height_width[1]
fov = float(cmd.get("field_of_view"))
radius = -cmd.get_view()[11]
height = radius * math.sin(math.pi * fov / 180.0)
width = height * asp_ratio
# translate side1 off to the right
cmd.translate([width,0,0],"side1")
# and rotate side2 by 180 degrees so that it faces us
cmd.rotate("y",180,"side2")
# then move the camera so as to be able to see both
cmd.move("x",-width/2)
cmd.move("z",-radius)
cmd.origin("center")
# hide the selection indicator
disable interf
# adjust the clipping parameters
clip atoms, 3, interf
set fog_start, 0.7
# set colors and representations
color white
color orange, interf
as surface
bg white
# and render
ray
figure/splay_interface.txt · Last modified: 2013/08/19 21:00 (external edit)