media:atomtyping
Table of Contents
Atom Typing
Quick Notes
- The atom_type field in PyMOL is called “text_type”
label *, text_type
- Multiple atom type formats are available. You can choose: mol2, sybyl, macromodel, and sdf. The default is MOL2.
set atom_type_format, X
- Types are also written to file with:
save ligand.mol2, ligand
Notes
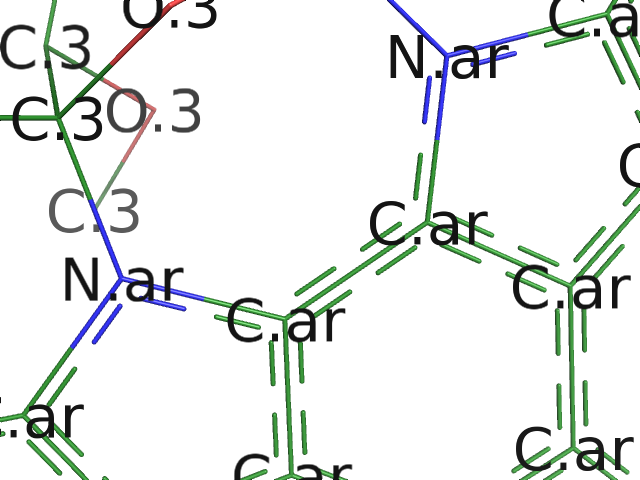 PyMOL can calculate, label, and write atoms types for molecules. Like stereochemistry, this happens at the level of the
PyMOL can calculate, label, and write atoms types for molecules. Like stereochemistry, this happens at the level of the object in PyMOL. (We plan to improve this for the next release.) Thus, for improved speed, it's beneficial to extract your ligand from a large protein before labeling it.
- Please note, you do not need to label the atoms before saving. PyMOL will read your desired file name extension (mol2, mmod) and type the atoms correctly before saving.
Settings
- atom_type_format (string, default: mol2) Sets the label format for label types. Supported options are: mol2, sybyl, macromodel, mmd, sdf.
- mol2
- sybyl (synonym for mol2)
- macromodel
- mmd (synonym for macromodel)
- sdf (default)
media/atomtyping.txt · Last modified: 2013/11/06 21:08 by vertrees

