Table of Contents
Stereochemistry in PyMOL
Quick Notes
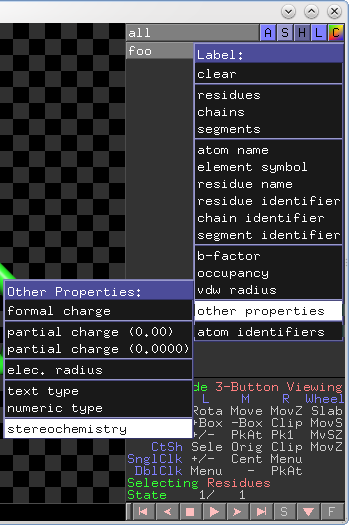
- L > Label > Other Properties > Stereochemistry
label ligand, stereoselect stereo “S”select stereo “R”select stereo “ ”color blue, stereo “R”
- Stereo does not work with alternate coordinates
- See
“help stereochemistry”
Details
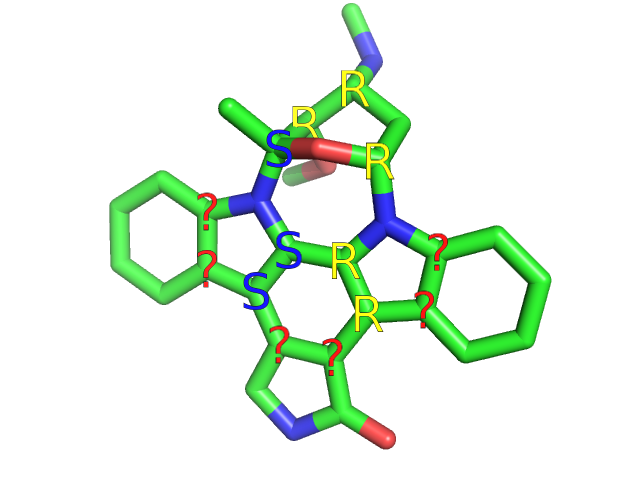
PyMOL v1.4 ships with the ability to calculate and show stereochemical labels. It uses a custom version of the MMSTEREO library from the Schrodinger software suite.
PyMOL adheres to the IUPAC nomenclature using the R/S system; therefore, labels will be “R”, “S”, “r”, “s”. (“r” and “s” are pseudoasymmetric labels.)
Nonchiral centers are labeled (and selected) with a blank string (in double quotes): “ ”. PyMOL labels atoms with a “?” when it realizes the atom is chiral, but for some reason (eg. geometry) cannot uniquely identify a label. If you find a molecule that PyMOL fails to label bit you think it should be able to label, please email help@schrodinger.com.
Chiral labels can be selected using the stereo selection modifier, eg
select stereo “R”
where the possible selections are: “R”, “S”, “r”, “s”, “ ” (blank).
Alternate Coordinates
Chirality is determined by the chemical and structural properties of non-identical liganded constituents. By nature of the CIP-rules, a change at one atom position could lead to a cascading effect of changes in other labels. Because of this, PyMOL will not label alternate coordinates. If we allowed alternate coordinates to be labeled, we would have to allow the determination of the alternately located atoms to change to reflect the alternate coordinates. But, the original determination was already correct, so there could be a confusion.
If you need to label an atom with alternate coordinates, you still can. But, you have to prepare the structure by hand. You have two options; here's how:
- If the structure has alternate coordinate labels, but only one atom is present in the structure then you can simply clear the alternate coordinate label(s) for that structure:
alter objectName, alt=“”
- If you have a structure with many alternate coordinates, you can select, by residues (or even atom), which coordinates you want. Here's are some examples:
# fetch a protein with alternate coordinates fetch 1oky # remove the ateernate coordinates remove alt "B" # clear the alternate coord. field in the structure alter 1oky, alt="" # label the entire protein label *, stereo
or
# fetch a protein with alternate coords fetch 1oky # create a new object without alternate coordinates "A" create altObj, 1oky and (not alt "A") # clear the alternate coord. field in the structure alter altObj, alt='' # label the new structure label altObj, stereo
Settings
set auto_defer_atom_count, X # any structure with < X atoms is automatically labeled upon loadingset auto_defer_atom_count, 0 # do not auto label anything